Add of new documentation pages + only doc build for ci for now
Showing
- .gitlab-ci.yml 1 addition, 1 deletion.gitlab-ci.yml
- docs/src/api/model.md 8 additions, 0 deletionsdocs/src/api/model.md
- docs/src/api/plots.md 8 additions, 0 deletionsdocs/src/api/plots.md
- docs/src/api/trajectory.md 8 additions, 0 deletionsdocs/src/api/trajectory.md
- docs/src/assets/logo.png 0 additions, 0 deletionsdocs/src/assets/logo.png
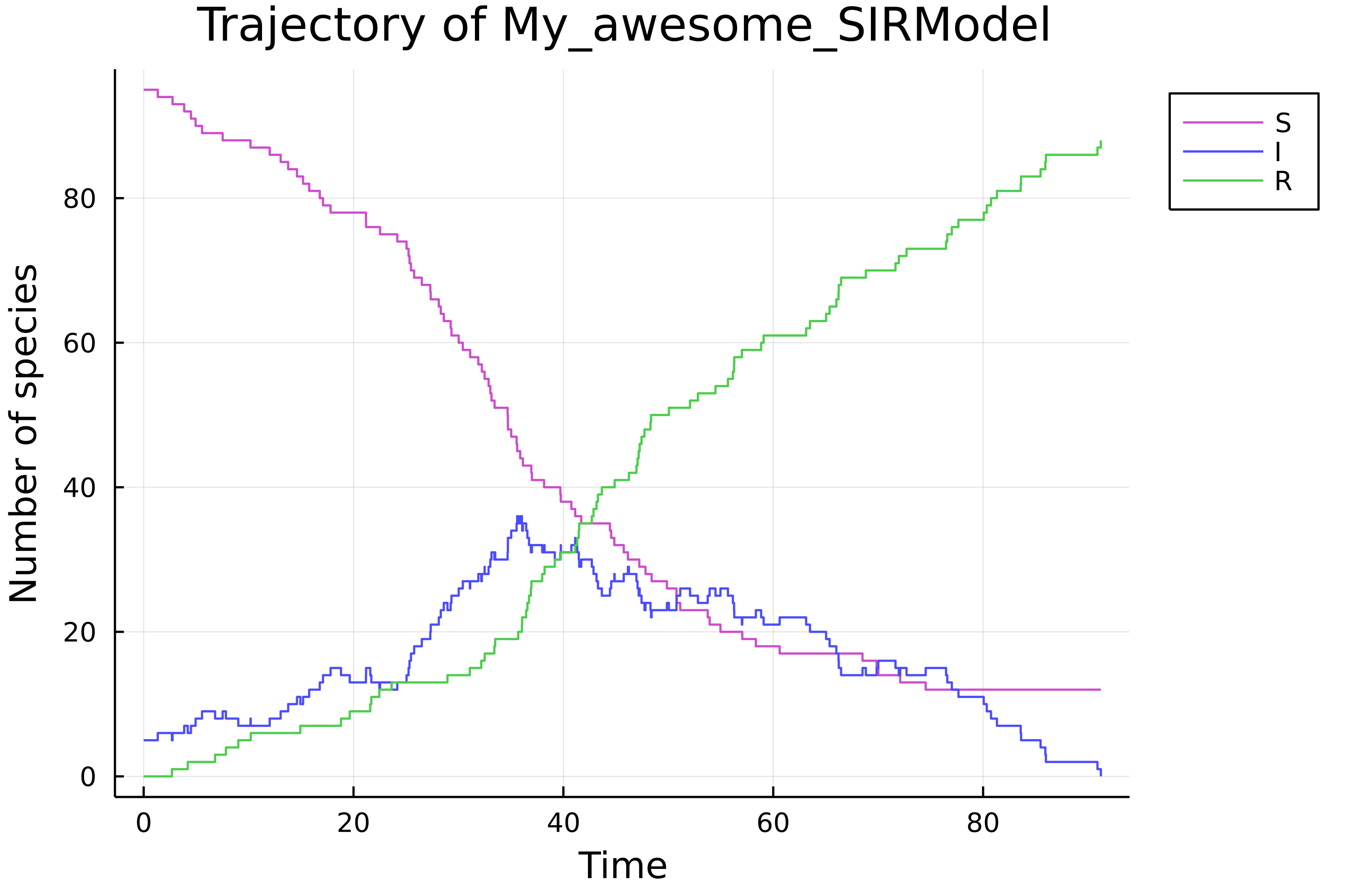
- docs/src/assets/sir_trajectory.png 0 additions, 0 deletionsdocs/src/assets/sir_trajectory.png
- docs/src/create_model.md 197 additions, 0 deletionsdocs/src/create_model.md
- docs/src/starting.md 127 additions, 0 deletionsdocs/src/starting.md
docs/src/api/model.md
0 → 100644
docs/src/api/plots.md
0 → 100644
docs/src/api/trajectory.md
0 → 100644
docs/src/assets/logo.png
0 → 100644
2.98 KiB
docs/src/assets/sir_trajectory.png
0 → 100644
132 KiB
docs/src/create_model.md
0 → 100644
docs/src/starting.md
0 → 100644